Dilute to Enrich for Deeper Proteomics
Dilute to Enrich for Deeper Proteomics: A Yolk-Depleted Carrier for Limited Populations of Embryonic (Frog) Cells
Leena R Pade, Camille Lombard-Banek, Jie Li, Peter Nemes
J Proteome Res. 2023 Nov 23. doi: 10.1021/acs.jproteome.3c00541.
Click here to view article at Journal of Proteome Research.
Click here to view article at PubMed.
Click here to view article on Xenbase.
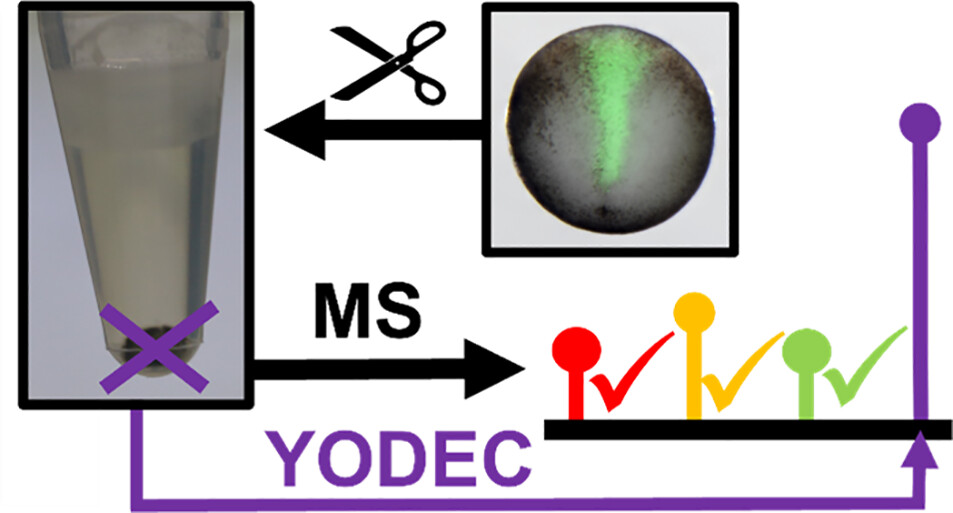
Abstract
Abundant proteins challenge deep mass spectrometry (MS) analysis of the proteome. Yolk, the source of food in many developing vertebrate embryos, complicates chemical separation and interferes with detection. We report here a strategy that enhances bottom-up proteomics in yolk-laden specimens by diluting the interferences using a yolk-depleted carrier (YODEC) proteome via isobaric multiplexing quantification. This method was tested on embryos of the South African Clawed Frog (Xenopus laevis), where a >90% yolk proteome content challenges deep proteomics. As a proof of concept, we isolated neural and epidermal fated cell clones from the embryo by dissection or fluorescence-activated cell sorting. Compared with the standard multiplexing carrier approach, YODEC more than doubled the detectable X. laevis proteome, identifying 5,218 proteins from D11 cell clones dissected from the embryo. Ca. ∼80% of the proteins were quantified without dropouts in any of the analytical channels. YODEC with high-pH fractionation quantified 3,133 proteins from ∼8,000 V11 cells that were sorted from ca. 2 embryos (1.5 μg total, or 150 ng yolk-free proteome), marking a 15-fold improvement in proteome coverage vs the standard proteomics approach. About 60% of these proteins were only quantifiable by YODEC, including molecular adaptors, transporters, translation, and transcription factors. While this study was tailored to limited populations of Xenopus cells, we anticipate the approach of "dilute to enrich" using a depleted carrier proteome to be adaptable to other biological models in which abundant proteins challenge deep MS proteomics.
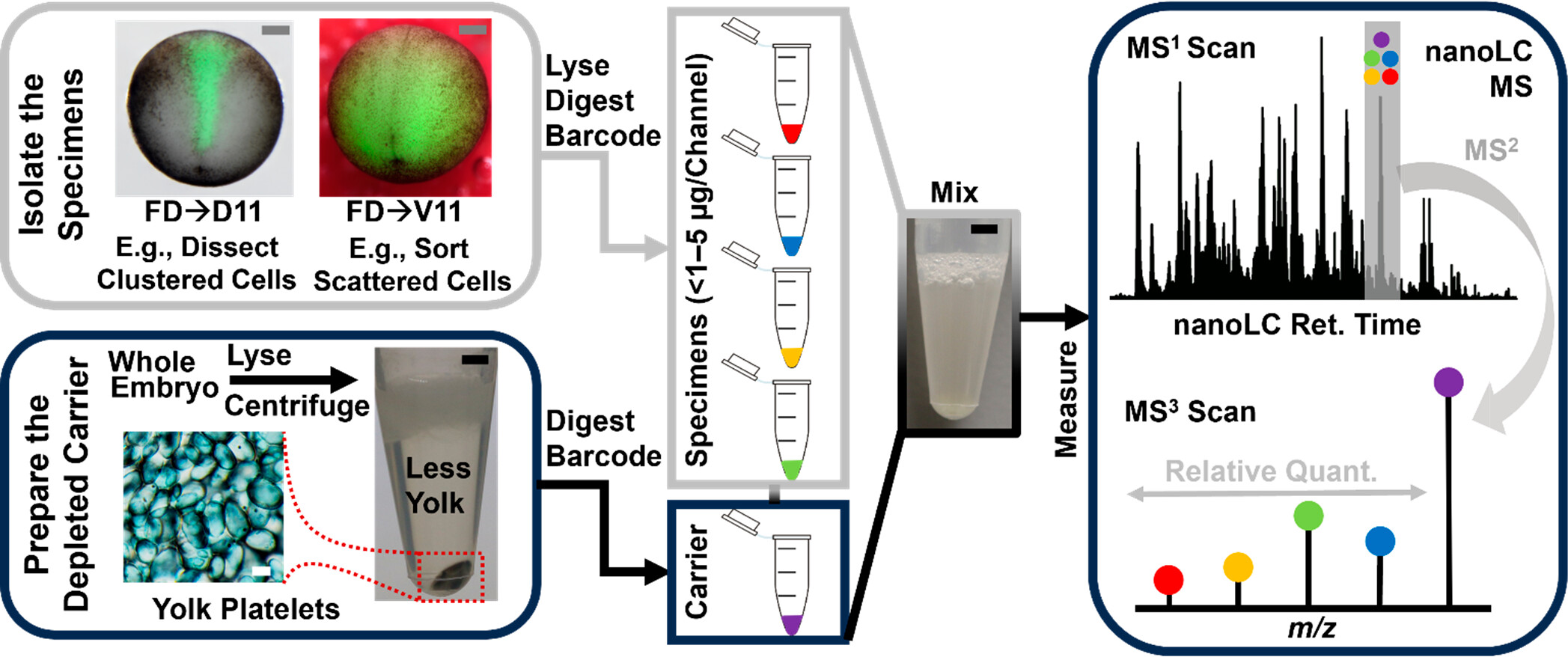
Figure 1. Our experimental strategy of “dilute to enrich” using a yolk-depleted (YD) carrier (YODEC). The example shows the fluorescent dextran (FD) labeling of identified neural-tissue-fated D11 and epidermal precursor V11 cell lineages in the X. laevis embryo (stage 13). The tight cluster of FD-labeled D11 cell lineage was dissected. The FD-labeled V11 clone, which scatters the embryo surface, was isolated through fluorescence-activated cell sorting (FACS). The collected proteomes were processed and TMT-tagged, then mixed with an isobarically tagged proteome digest prepared from a YD proteome. The samples were analyzed using nanoLC-HRMS employing multinotch precursor isolation and MS3-level quantification. Scale bars, 250 μm (gray); 2 mm (black).
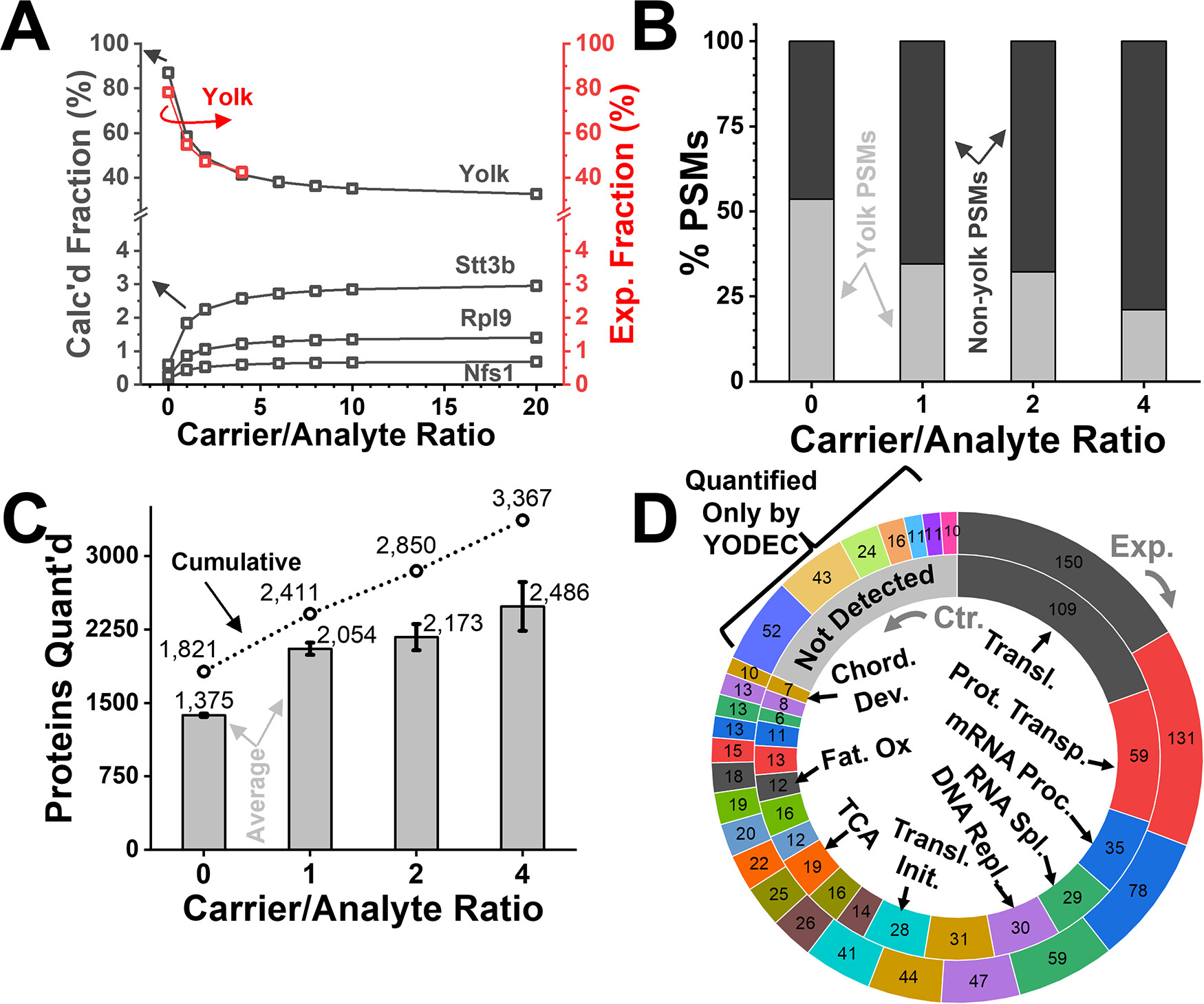
Figure 3. YODEC improves the proteome coverage and quantification. (A) Fractional composition of the X. laevis proteome upon progressive dilution with the yolk-depleted proteome from in silico modeling (black) vs experimental testing (red). For a proteome mixture containing TMT-barcoded X. laevis proteome at a 1.5:1:1:1.5 analytical ratio, increasing carrier proteomes (B) reduced PSMs on yolk peptides and (C) improved the number of quantifiable proteins. (D) Comparison of gene ontology assessment of molecular function for proteins that were quantified without (Control, inner circle) and with YODEC using a 4-fold carrier load (Experimental, outer circle). Close-up in Figure S5A. Identical GO categories are coded in the same color between the control (inner circle) and experimental (outer circle) cases.
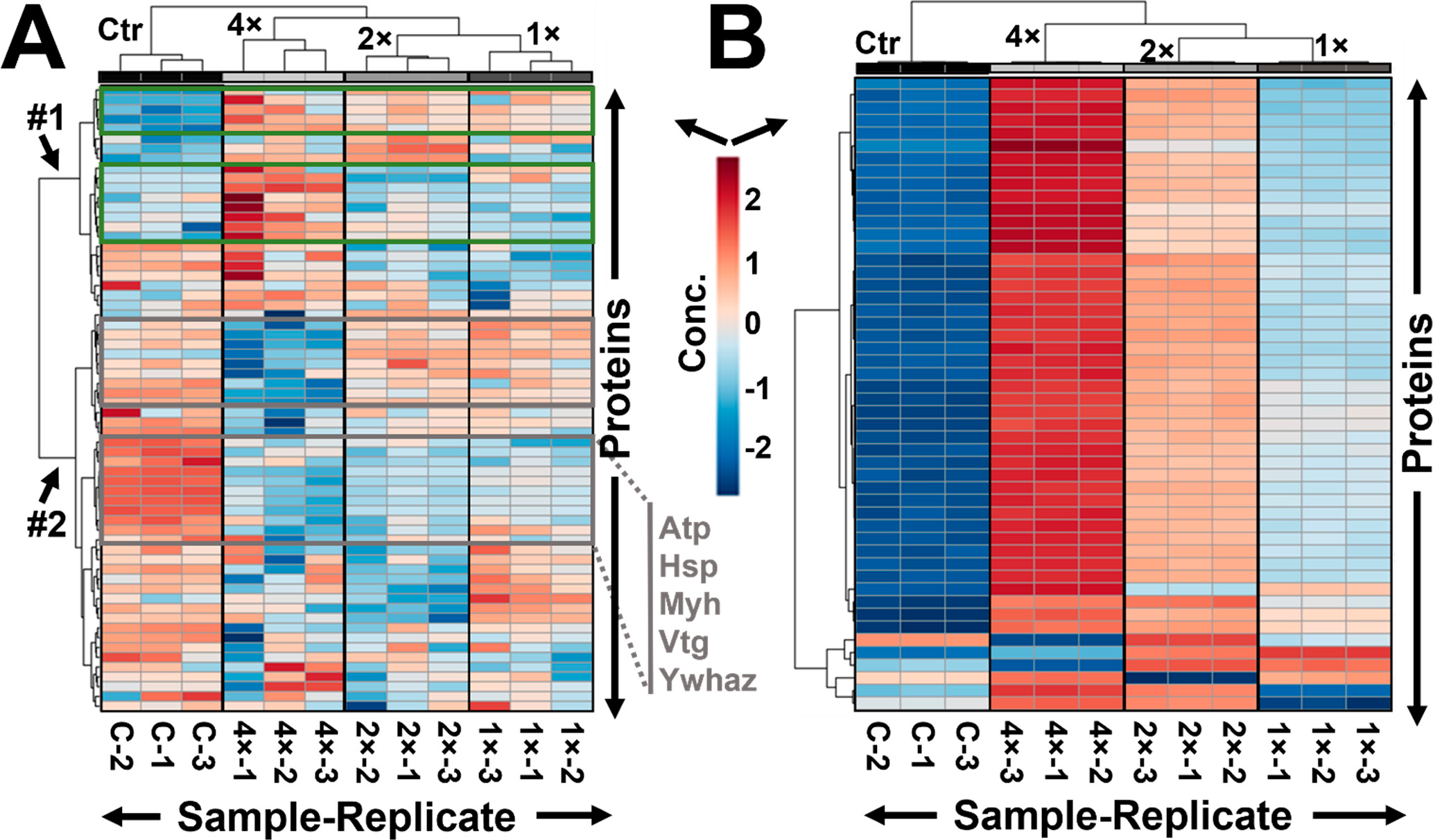
Figure 4. Mechanistic comparison of the “dilute to enrich” approach using the X. laevis and HeLa proteomes. Heatmap and unsupervised cluster analyses on the 50 statistically most significantly differential proteins between the control (C) and experimental conditions using carrier proteomes in 1-, 2-, and 4-times excess (samples labeled 1×, 2×, and 4×, respectively). Each sample was analyzed in three technical replicates (sample labels appended with −1, −2, or −3). These measurements revealed (A) differential proteome enrichment during YODEC, where the X. laevis proteome was mixed with its yolk-depleted proteome (yielding Groups #1 and #2), (B) but not in the carrier proteome (SCoPE) strategy on HeLa, where protein amounts were uniformly increased. Close-ups of these plots are available in Figure S6b. Key to color code: blue, signal loss; red, signal enrichment. Sample labels specify the experiment (control vs YODEC carrier) and the number of the technical replicate.
Adapted with permission from ACS Publications on behalf of Journal of Proteome Research: Pade et al. (2023). Dilute to Enrich for Deeper Proteomics: A Yolk-Depleted Carrier for Limited Populations of Embryonic (Frog) Cells. J Proteome Res. 2023 Nov 23. doi: 10.1021/acs.jproteome.3c00541.
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
